The significant importance of the detection of pathogens, I will assume, is perfectly clear to anyone reading this blog. However, I believe it may make sense to discuss some of the general science that permits the declaration of foods as either safe or contaminated.
Throughout my long, long career in bioanalytical science, I always felt like a detective trying to solve a case. Indeed, we have a lot in common with forensic scientists. We both search for “suspects”; we both need to identify those suspects with 100% confidence; and the price of being wrong could be pretty high in both cases as well.
However, the major difference is, forensics uses a known crime scene to then hunt for suspects, while we hunt for suspects without knowing if a “crime” has even been committed. Methods used to identify suspects are another area in which we must use slightly different techniques, based on cost, time and confidence level.
For example, a photo that implicates a suspect seems to offer a high level of confidence of proper identification. Yet, the photo needs to have been taken at a good resolution, the correct angle, etc. Otherwise, proper identification of the suspect cannot happen with 100 percent confidence. Forensics might increase the confidence level of suspect ID by matching fingerprints or DNA at the crime scene. In some ways, we do the same at our “crime scenes”! Yes, microbes don’t leave fingerprints (due to a lack of fingers!), however, microbes do have DNA. So, just like our forensics brethren, we can use a most reliable way to find and identify our suspect.
What is DNA?
Again, most anyone reading this blog post know the basics on what DNA is, but fewer people know the basics on its structure.
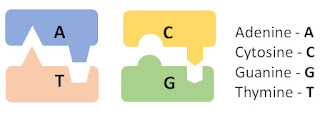
DNA molecules are built of just four nucleobases: Adenine (A), Cytosine (C), Guanine (G), and Thymine (T). These four nucleobases code all genetic information through their sequence in a chain, in similar fashion to the way that computers code all information via sequences of “on” and “off” switches — at a very basic level. But it is made more complicated by the fact that DNA forms not just one but two chains that are connected two each other, forming a double helix.
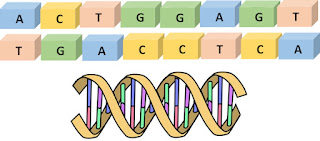
The double helix is not just two random chains, however, because the chains can bind to one another only where A binds to T and C binds to G.
DNA as an identification tool
DNA is a convenient object for pathogen detection
based on some important properties of this substance. First, with some
manipulations, DNA can form millions of copies out of one molecule. Moreover,
one does not need copies of the whole molecules. One needs only copies of a
part of DNA that codes a unique feature of an organism. For example, to detect
Shiga toxin-producing E. coli (STEC), one needs to identify only small
parts of DNA unique for coding E. coli itself and for coding a gene
responsible for Shiga toxin. The rest of DNA molecule is not really needed.
The ability to create numerous copies of molecule
fragments out of one molecule (known as “amplification”) is an extremely
powerful tool as well, in that it solves the issue of sensitivity in a great
way. Most DNA-based detection methods start with amplification through a method
called Polymerase Chain Reaction (PCR).
PCR: how it works
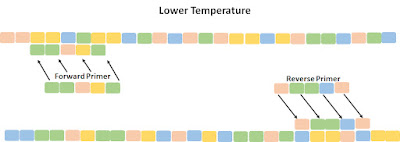
PCR-based amplification employs a basic unit called a thermocycler. The machine can very quickly alternate the temperature of a low-volume reaction medium. A cycle starts by increasing the temperature to 95-98 C. Under these conditions, the two strands of the suspect double helix break apart. The temperature then is quickly lowered, allowing the DNA strands to rebind.
However, primers — short, chemically synthesized portions of DNA molecule that perfectly match the end of the “suspect” DNA fragment of interest — have been added to the solution in great numbers (think, billions of primers), so the chains typically rebind with two primers. Because of the extreme population of primers in the solution, there is a much higher chance the DNA strands will bind to primers rather than its original partner.
Along with primers, the reaction medium contains “building material” to complete the new double helixes: A, C, G, T, and polymerase enzyme. Primers binding to the chains trigger a polymerase chain reaction.
This reaction occurs only in the presence of a primer specifically attached to a chain. So, the second chain is quickly built over each of thermally separated chains. This forms two complete copies of the original double helix, doubling DNA material and creating two molecules out of one. The cycle restarts with the heating of the solution, and four molecular copies are made. Each subsequent cycle doubles number of copies.
We now have many millions of copies of DNA; not just any copies, but importantly, copies of the small DNA fragment that we are interested in. Proper selection of the primer pairs allows us to pick up only the part of DNA of interest, and amplification occurs only for that part of the molecule between forward and reversed primers. Thus, we narrowed down our area of interest and created a huge number of identical DNA pieces for further investigation.
How difficult is to run PCR?
Running PCR is an easy task nowadays, though it was a
challenge to develop. A user receives a kit with everything that is needed in a
tube. Users simply add a DNA sample into the tube, close the tube, set it into
a PCR thermocycler, and press a start button. The process is usually completed
in about one hour.
Does PCR always work properly?
Just like any process, PCR is not perfect: sometimes it fails. There are a variety of reasons why amplification might not occur or the wrong molecules were amplified, including simple operator error. However, PCR generally is a very robust, reliable technique with very low rate of failure.
What happens when PCR is done?
The final PCR product (called the amplicon) provides a
fair amount of molecules for subsequent pathogen-detection steps, since PCR
alone does not yet answer a question on presence/absence of a pathogen DNA in
the sample. PCR simply provides the amplicon to more accurately determine a
pathogen’s presence.
There are multiple methods — some more
complicated, others less so — to answer this main question. Some
only identify the presence/absence of any amplicon in PCR product. Certainly,
if there is no amplicon, there is no pathogen in the sample. But these
techniques may provide false positive data, particularly if the PCR process
amplified the wrong molecules. Other techniques provide data on a structure of
amplified molecule, and thus are pretty robust in terms of a rate false
positives.
Nevertheless, when you’re looking for pathogen
“suspects” via DNA-based methods, PCR amplification will be at the core of the
process you’re going to use, and after reading this post, you will hopefully
have a better grasp on how PCR works.
— Andrei Gindilis, senior scientist, andrei@werfoodsafety.com